The Mechanism of Voltage Gating in Potassium Channels? – Part 1 Deactivation
In this two part post I will discuss the recent Science paper (April 13th issue) from the D.E. Shaw Research group, entitled “The Mechanism of Voltage Gating in Potassium Channels.” In this paper Jensen et al. use their custom designed supercomputer Anton (named after Anton von Leeuwenhoek, the 18th century microscopist) to perform molecular dynamics (MD) simulations of voltage-gated K+ channels under the influence of different membrane potentials. As their starting point they used the structure of the Kv1.2-2.1 paddle chimera solved by Long et al. in 2007 to 2.4 Å resolution (PDB accession code 2R9R) (Long et al., 2007). This structure is of the channel in the activated (open) conformation which occurs during depolarization of the cell membrane (positive membrane potentials). Jensen et al. want to see if they can use Anton to calculate the deactivated (closed) state of the channel through MD simulations.
I work on a voltage-gated protein (Hv) and I have spent some time thinking about and studying the mechanism of how these voltage sensors operate to open and close the pore of the ion conduction pathway. So I have to say that watching the movies of these proteins in action generated from the MD simulations is pretty cool! However, just because something is cool doesn’t mean it is correct, so in this post I will try to overcome the coolness factor and be critical, and really look at what we might be able to learn from these simulations.
Jensen et al.‘s paper is filled with complex diagrams tracking the movements of specific residues of interest within the voltage-sensor domain (VSD) and pore domain structures. All of these figures are generated from the 15 simulations discussed in the paper, representatives of which are shown in the supplementary movies on-line (Jensen et al., 2012 supplementary). I recommend checking out the paper and having a look at the figures since they highlight some of the most important conformational changes; here I will mostly be using the movies themselves to discuss their results.
—
Deactivation – Closing the channel
The supplementary movies demonstrating the deactivation of the channel are from simulation number 9 in the paper (except movie S5 which is from simulation 5, see below). In simulation 9 the starting structure is the crystal structure of the paddle chimera (Long et al., 2007). The total length of the simulation was 256 microseconds (μs). During the simulation the protein is allowed to sit at 0 mV for ~2 μs after which the membrane potential was ramped over 1 μs to -350 mV held constant until ~205 μs where it was then increased to -500 mV (Jensen et al., 2012). This is a huge hyperpolarization! In reality, the hyperpolarized membrane potential of a cell is no greater than -120 mV and the resting potential is usually around -70 mV. If you were doing an electrophysiological experiment (i.e. a patch clamp experiment), stepping to this extreme voltage would break the membrane seal. The necessity for these extreme, non-physiological membrane potentials in the simulations suggests to me that the force fields used in the MD calculations have some systematic flaw.
The physics of molecular force fields are not my area of expertise, but when you look more closely at the supplementary methods you start to get an idea for what that flaw may be (Jensen et al., 2012 supplementary). The force fields used in the simulations were: CHARMM27, for protein, ions and water; and CHARMM36, for lipids (MacKerell et al., 1998; Kluada et al., 2010). However, using the standard parameters in the simulations they were unable to see any movement of the voltage sensors, even when using the extreme hyperpolarizing membrane potential of -750 mV (simulation 8 first 45 μs). However, the authors were able to get the voltage-sensors to move by weakening the guanidinium acetate association constant (Jensen et al., 2012 supplementary). This “DER” (Aspartate-Glutamate-Arginine) correction reduced the 4.6 M-1 association constant between Asp-Arg or Glu-Arg salt-bridging interactions obtained using CHARMM27 to 0.9 M-1 (DER) or 0.4 M-1 (DER2) which agrees better with experimental values of 0.3-0.5 M-1 (Springs and Haake, 1977). CHARMM27 was off by an order of magnitude! It makes sense that overly strong salt bridging interactions between these residues would prevent voltage-sensor movement since the structure contains several such interactions between the S4 arginines and aspartates or glutamates on S1 and S2 (Long et al., 2007). Does CHARMM27 alway over estimate this interaction strength or is it only because of the low dielectric environment of the membrane? Since I am not an expert in the use of these force fields, and I suspect that most electrophysiologists who have been interested in and studying this process for decades are also unfamiliar with the finer points of CHARMM27, I would have really appreciated the authors spending more time discussing the reasons for the necessity of these corrections.
Simulation 9 (the representative simulation in used in movies S1, S2, S3 and S4) was started using the DER correction, but then at 78 μs it was switched to the DER2 correction. The main difference between the two being a significant decrease in the partial charge of the arginine side chain (Jensen et al., 2012 supplementary). Only after the switch to DER2 did the voltage-sensors undergo the conformational change. Even with the DER2 correction the non-physiological hyperpolarization of -350 mV was still required for movement of the voltage-sensors (Jensen et al., 2012 supplementary). This indicates to me that other parameters of CHARMM27 may generate additional overly strong interactions.
Ok, so there are a few caveats: the membrane potentials are extreme and in reality the charge of an aginine side chain doesn’t spontaneously decrease during a hyperpolarization. But let’s check out the movies and see what happens during the induced voltage-sensor conformational changes.
Movie S1 & Movie S2 from Simulation 9, T1 domain present
These are the intracellular (movie S1) and extracellular (movie S2) views of deactivation simulation 9. If you don’t immediately recognize what you are looking at, there is a description of the coloring scheme in the supplementary information that should help orient you (Jensen et al., 2012 supplementary). There are two main features that these movies emphasize: the closing of the intracellular gate (blue residues in the center) and the disassociation of the voltage-sensor domains (green and red helices) from the pore (white helices). In movie S1 you see the pore start to close at ~35 μs (timer on top left) and finish closing completely at ~130 μs. It is interesting to note that although the channel is a four-fold symetric tetramer, the closing of the channel presented here is not four-fold symmetric. This symmetry breaking during closure was also seen in the groups earlier simulations of the isolated Kv1.2 pore domain in the absence of VSDs (Jensen et al., 2010). It is certainly true that proteins are more than capable of this sort of symmetry breaking, however the closed structures of the bacterial K+ channel KcsA and the eukaryotic Kir2.2 and Girk2 channels are all four-fold symmetric (Zhou et al., 2001; Tao et al., 2009; Whorton et al., 2011). The only case that I am aware of for a non four-fold symmetric closed structure of a K+ channel is of a mutant of Girk2 with only two of the four protomers binding to PIP2 (Whorton et al., 2011; PDB accession code 3SYQ).
Movie S5 from Simulation 5, T1 domain absent
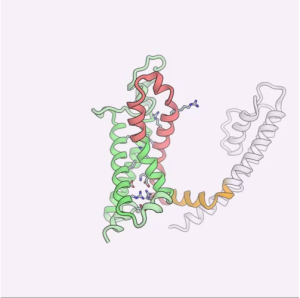 The other interesting feature is the disassociation of the VSDs from the pore which can be observed in both movies. Most drastically at ~130 μs, just as the pore completes it’s closure, one of the VSDs disassociates and swings about (movie S1: top left VSD, movie S2: bottom left VSD). This VSD dissociation is more pronounced in movie S5 (simulation 5), which was performed in the absence of the intracellular T1 domain. This 150 μs simulation was performed with the DER correction with initial ramp depolarization to -750 mV, the voltage was then decreased to -375 mV after all VSDs had undergone conformational change at ~70 μs (Jensen et al., 2012 supplementary). In the movie, the conformational change in S4 happens very quickly (I will discuss the movement of the S4 arginines more below) then the VSD completely swings out away from the pore domain and the S4-S5 linker (yellow helix) is stretched.
The other interesting feature is the disassociation of the VSDs from the pore which can be observed in both movies. Most drastically at ~130 μs, just as the pore completes it’s closure, one of the VSDs disassociates and swings about (movie S1: top left VSD, movie S2: bottom left VSD). This VSD dissociation is more pronounced in movie S5 (simulation 5), which was performed in the absence of the intracellular T1 domain. This 150 μs simulation was performed with the DER correction with initial ramp depolarization to -750 mV, the voltage was then decreased to -375 mV after all VSDs had undergone conformational change at ~70 μs (Jensen et al., 2012 supplementary). In the movie, the conformational change in S4 happens very quickly (I will discuss the movement of the S4 arginines more below) then the VSD completely swings out away from the pore domain and the S4-S5 linker (yellow helix) is stretched.
It has been shown that the VSD makes two functionally significant contacts with the pore domain: one via the S4-S5 linker and the other between S1 and the pore helix (Lee et al., 2009). Interestingly, when a disulfide cross-bridge was engineered across the S1 and pore interface in the archeal voltage-gated K+ channel KvAP, the channels where still functional and voltage-gated. However, unlike wild type KvAP, which is a very strongly inactivating channel (Ruta et al., 2003; Schmidt et al., 2009), the currents from the cross-bridged channels did not inactivate (Lee et al., 2009). This suggests to me that disassociation of the VSD and pore may be indicative of an inactivated state and is not an essential part of deactivation.
The larger dissociation from the pore domain in the absence of the T1 domain (simulation 5) versus in the presence of the T1 domain (simulation 9) led the authors to propose that a functional role of the T1 domain may be to restrain the VSD, keeping it close to the pore domain. Jensen et al. point out that this result agrees with experiments which show that in the absence of a T1 domain the channels activate with slower kinetics (Kobertz et al., 1999). The further away the VSDs get from the pore, the longer it takes for them to reassociate with the pore and open the channel. However, in the experiments by Kobertz et al., the removal of the T1 domain also reduces the rate of channel deactivation (closing) significantly. In all the deactivation simulations performed in the absence of T1 (simulations 5-8), the closing of the channel was significantly faster than simulation 9 in the presence of T1 (Jensen et al., 2012 supplementary). This discrepancy is not discussed in the paper.
The really cool features of the deactivation simulations are those that happen in the VSD themselves, so let’s have a look at some more movies.
Movie S3& S4 from Simulation 9, T1 domain present
Movie S6 from Simulation 9, T1 domain present
These movies (and movie S5 of simulation 5) all focus on the internal conformational changes inside the VSD. The major movement is of the S4 helix in the VSD. It is well established that the positively-charged amino acid residues on S4 carry the gating-charges and undergo a conformational change in response to changes in membrane potential thereby opening and closing the channel (Aggarwal & MacKinnon, 1996). It has also been shown that the conformational changes in S4 occur as a series of discrete transitions (Sigg et al., 1994). Furthermore, we know these transitions are centered around a charge transfer center formed by Phe233, Glu236 and Asp259 in the middle of the VSD (Tao et al., 2010). It has been shown, through direct measurement of gating charge movements, that by mutating this site it is possible to trap the VSD in different conformations that correspond to different gating-charge residues on the S4 helix (the arginines and lysine highlighted in the movies) occupying this site (Tao et al., 2010). Basically, it is known that in order to go from the activated (open) to deactivated (closed) state (or vice versa) of the VSD, these charged amino acid residues pass into and out of this charge transfer center in the middle of the membrane. The coolest part of these simulations is getting to see this process explicitly. For example, in movie S6 you see the first gating charge transition at ~80 μs and then another two transitions in quick succession at ~240 μs.
These simulations also show that, as the S4 charges pass through the charge transfer center the helix rotates about its axis, generating a screw-like motion. This helical-screw model of S4 motion has also been previously predicted (Tombola et al. 2005; Tombola et al., 2006; Campos et al., 2007; Broomand and Elinder, 2008). However, none of the studies that predicted the S4 rotation had directly measured rotation of S4. The experimenters inferred rotation by measuring the positions of different residues along S4 relative to other residues in the VSD, in both the open and closed states. The only issue I have with this line of argument is that in the high-resolution structure of the paddle chimera the top half of S4 is not an α-helix but a 310 helix (Long et al., 2007). This has profound implications for the helical-screw model, since the S4 gating charge residues, which undergo the above mentioned conformational change, occur along S4 every third residue. Essentially, this means that, if S4 maintains its 310 helical structure, no rotation would have to occur at all for each of the gating charges to sequentially occupy the charge transfer site. The helical-screw model would have to become the sliding-helix model. I am not claiming that the helical-screw model is definitely wrong. It is certainly possible that S4 undergoes a 310-helix to α-helix transition during deactivation. However, I do find it very strange that Jensen et al. do not mention the S4 310-helix at all. I assume it is included in their starting model since they say they are using the paddle chimera structure. Does the 310-helix relax to an α-helix during the ~2 μs equilibration at 0 mV? I am genuinely interested to know what happened.
—
In my next post I will discuss simulations 10-14 in which Jensen et al. apply strong depolarizations to their calculated deactivated structures in order to try to open the channel back up. I will also share my general thoughts and opinions concerning the models generated.
—
Works Cited in this Post
Aggarwal, S. K., & Mackinnon, R. (1996). Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron, 16(6), 1169–1177.
Broomand, A., & Elinder, F. (2008). Large-Scale Movement within the Voltage-Sensor Paddle of a Potassium Channel—Support for a Helical-Screw Motion. Neuron, 59(5), 770–777. doi:10.1016/j.neuron.2008.07.008
Campos, F. V., Chanda, B., Roux, B., & Bezanilla, F. (2007). Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proceedings of the National Academy of Sciences of the United States of America, 104(19), 7904–7909. doi:10.1073/pnas.0702638104
Jensen, M. Ø., Borhani, D. W., Lindorff-Larsen, K., Maragakis, P., Jogini, V., Eastwood, M. P., Dror, R. O., et al. (2010). Principles of conduction and hydrophobic gating in K+ channels. Proceedings of the National Academy of Sciences of the United States of America, 107(13), 5833–5838. doi:10.1073/pnas.0911691107
Jensen, M. Ø., Jogini, V., Borhani, D. W., Leffler, A. E., Dror, R. O., & Shaw, D. E. (2012). Mechanism of voltage gating in potassium channels. Science, 336(6078), 229–233. doi:10.1126/science.1216533
Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T., & Mackinnon, R. (2002). Crystal structure and mechanism of a calcium-gated potassium channel. Nature, 417(6888), 515–522. doi:10.1038/417515a
Jiang, Y., Lee, A., Chen, J., Ruta, V., Cadene, M., Chait, B. T., & Mackinnon, R. (2003). X-ray structure of a voltage-dependent K+ channel. Nature, 423(6935), 33–41. doi:10.1038/nature01580
Klauda, J. B., Venable, R. M., Freites, J. A., O’Connor, J. W., Tobias, D. J., Mondragon-Ramirez, C., Vorobyov, I., et al. (2010). Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. The journal of physical chemistry B, 114(23), 7830–7843. doi:10.1021/jp101759q
Kobertz, W. R., & Miller, C. (1999). K+ channels lacking the “tetramerization” domain: implications for pore structure. Nature structural biology, 6(12), 1122–1125. doi:10.1038/70061
Lee, S.-Y., Banerjee, A., & Mackinnon, R. (2009). Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K(+) channels. PLoS biology, 7(3), e47. doi:10.1371/journal.pbio.1000047
Long, S. B., Campbell, E. B., & Mackinnon, R. (2005). Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science, 309(5736), 897–903. doi:10.1126/science.1116269
Long, S. B., Tao, X., Campbell, E. B., & Mackinnon, R. (2007). Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature, 450(7168), 376–382. doi:10.1038/nature06265
MacKerell, A. D., Bashford, D., Bellott, Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., et al. (1998). All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins †. The journal of physical chemistry B, 102(18), 3586–3616. doi:10.1021/jp973084f
Ruta, V., Jiang, Y., Lee, A., Chen, J., & Mackinnon, R. (2003). Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature, 422(6928), 180–185. doi:10.1038/nature01473
Schmidt, D., Cross, S. R., & Mackinnon, R. (2009). A gating model for the archeal voltage-dependent K(+) channel KvAP in DPhPC and POPE:POPG decane lipid bilayers. Journal of Molecular Biology, 390(5), 902–912. doi:10.1016/j.jmb.2009.05.062
Sigg, D., Stefani, E., & Bezanilla, F. (1994). Gating current noise produced by elementary transitions in Shaker potassium channels. Science, 264(5158), 578–582.
Springs, B., & Haake, P. (1977). Equilibrium constants for association of guanidinium and ammonium ions with oxyanions. Bioorganic Chemistry, 6(2), 181–190. doi:10.1016/0045-2068(77)90019-0
Tao, X., Avalos, J. L., Chen, J., & Mackinnon, R. (2009). Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science, 326(5960), 1668–1674. doi:10.1126/science.1180310
Tao, X., Lee, A., Limapichat, W., Dougherty, D. A., & Mackinnon, R. (2010). A gating charge transfer center in voltage sensors. Science, 328(5974), 67–73. doi:10.1126/science.1185954
Tombola, F., Pathak, M. M., & Isacoff, E. Y. (2005). Voltage-Sensing Arginines in a Potassium Channel Permeate and Occlude Cation-Selective Pores. Neuron, 45(3), 379–388. doi:10.1016/j.neuron.2004.12.047
Tombola, F., Pathak, M. M., Gorostiza, P., & Isacoff, E. Y. (2006). The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature, 445(7127), 546–549. doi:10.1038/nature05396
Whorton, M. R., & Mackinnon, R. (2011). Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell, 147(1), 199–208. doi:10.1016/j.cell.2011.07.046
Zhou, Y., Morais-Cabral, J. H., Kaufman, A., & Mackinnon, R. (2001). Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature, 414(6859), 43–48. doi:10.1038/35102009





Comments
2 Responses to “The Mechanism of Voltage Gating in Potassium Channels? – Part 1 Deactivation”Trackbacks
Check out what others are saying...[…] pass through this center “one by one” (for additional explanation and discussion, please see my earlier post and watch Jensen et al.’s simulations of the process). Is it possible that different VSDs open […]
[…] model in some detail while reviewing the work of Jensen et al. in previous posts (check it out here) and I will most definitely be writing more about the charge transfer center model of […]